The Gerlitz Lab
A. Chromatin Organization during Tumor Cell Migration
We have shown that induction of directed migration of melanoma cells is associated with and dependent on a global increase in chromatin condensation.
Induction of migration leads to an increase in various markers of heterochromatin formation: 1. Histone modifications associated with heterochromatin formation; H3K9me3, H3K27me3 and H4K20me1. 2. Increased chromatin residence time of histone H1 and decreased chromatin residence time of HMG proteins. 3. Increased DNA methylation. 4. Increased resistance to DNA degradation by DNase I.
Inhibition of heterochromatin formation by chemical inhibitors to histone methyl-transferases and HDACs as well as interfering with histone H1 function by a dominant negative form inhibits the cellular migration rate.
In looking for mechanistic roles of heterochromatin in migrating cells we mapped the genomic distribution of the heterochromatin markers H3K9me3, H3K27me3 and H4K20me1 following induction of migration by ChIP-seq. This analysis revealed migration-associated spreading out of these markers over larger genomic regions. This expansion is nonuniform; H3K9me3 and H4K20me1 are shifted towards repetitive elements whereas H3K27me3 is shifted towards genes.
Transcriptome analysis found out that H3K27 methylation is required for 33% of the migration-associated transcriptional changes. H3K27 methylation is also required to prevent transcriptional changes in a few hundred genes that are not supposed to change upon induction of migration. The most significantly affected pathway by H3K27 methylation is the cholesterol biosynthesis pathway.
Induction of B16F1 migration in the wound healing assay leads to an increase in H3K9me3 levels and the recognition of histone H1 by the AE4 antibody.
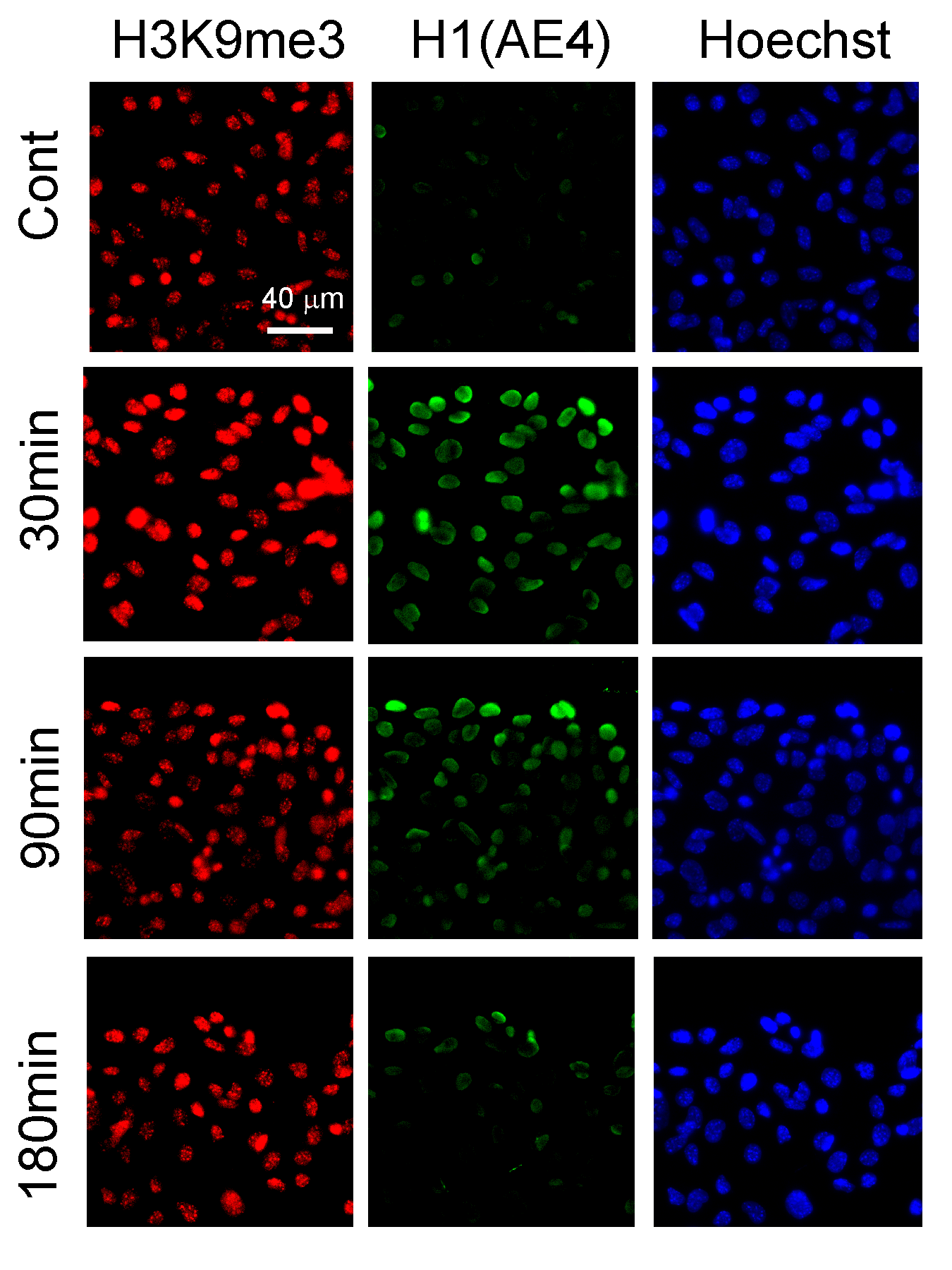
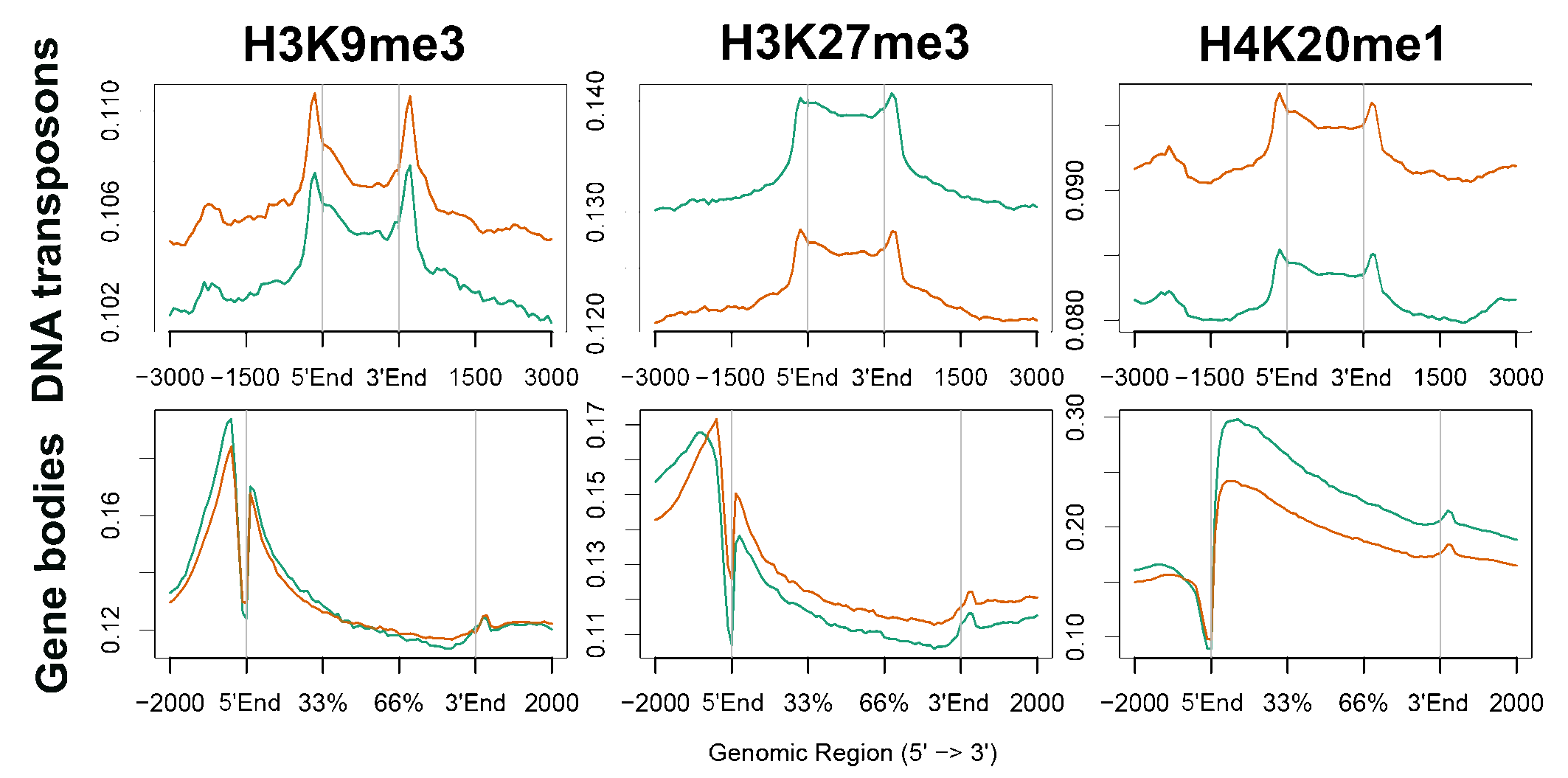
ChIP-seq signals distribution over transposones and genes in control cells and cells that were iduced to migrate for 3h.
B. Interrelations between Microtubules and Chromatin Organization
1. The microtubule regulation activity of the histone methyltransferase SETDB1:
The oncogene SETDB1 is a well-established histone methyltransferase that methylates H3K9 to induce heterochromatin and to alter transcription. Recently we found that a fraction of SETDB1 is associated with microtubules. SETDB1 promotes microtubule destabilization in a manner that is independent of its methyltransferase activity. SETDB1 appears to affect acetylation of alpha tubulin at Lys40, a modification that increases microtubule stability. SETDB1 associates with HDAC6, the major tubulin deacetylase, thus SETDB1 may promote HDAC6 activity.
2. Microtubule polymerization induces global chromatin alterations:
The ability of microtubule-generated forces to alter chromatin structure and organization during mitosis and meiosis is well documented. During interphase, interrelations between microtubules and the chromatin fibers were not expected to occur, since the microtubules are excluded from the metazoans’ nucleus by the nuclear envelope that forms a physical barrier between the chromatin fibers and the cytoplasm-localized microtubules. However, recently it has become apparent that microtubule-generated forces in the cytoplasm can affect chromatin dynamics inside the nucleus.
We were the first to show that synchronized microtubule polymerization in mammals is able to induce transient indentations in the nuclear envelope along with heterochromatin accumulation at the nuclear side that is the closest to the centrosome. The ability of the microtubules in the cytoplasm to alter heterochromatin organization inside the nucleus is dependent on the motor protein dynein and the composition of the nuclear lamina.
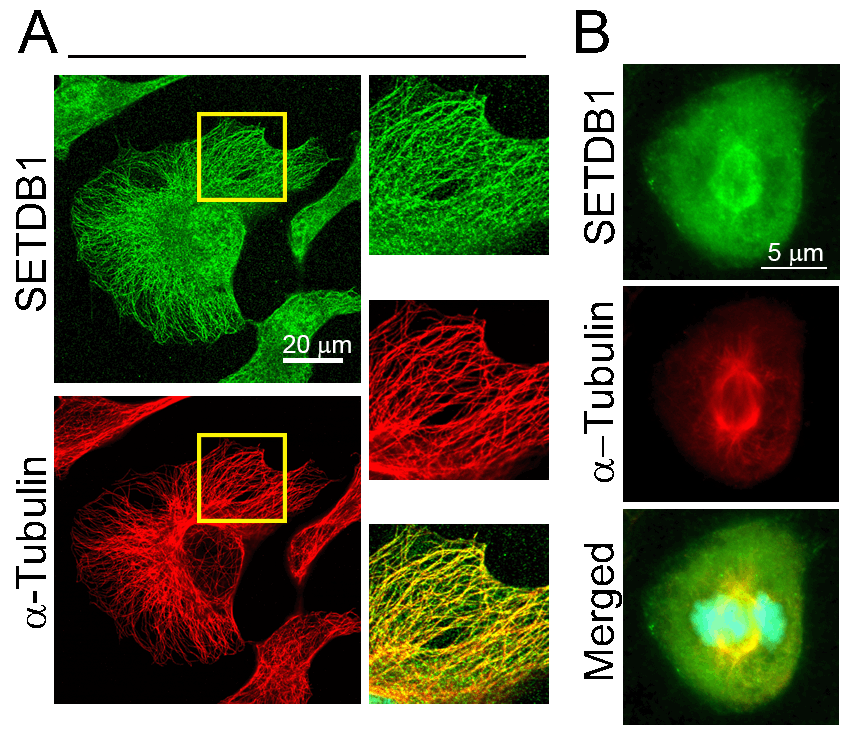
Colocalization of SETDB1 with microtubules in HeLa cells during interphase (A) and mitosis (B).

Microtubule recovery from nocodazole treatment induces indentations in the nuclear envelope along with heterochromatin accumulation at the nuclear side closest to the centrosome. Red- alpha tubulin, Yellow- gamma tubulin, Green- histone H1.